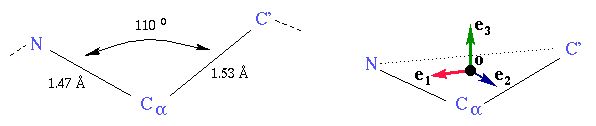
Non-related proteins can have very different structures, but when they share a common biological function, often they share a common 3D substructure that realizes this function: this is a motif. Binding motifs can be widely distributed in the protein sequence and involve only few, if any, consecutive amino acids. Thus we cannot really use the sequence and we have to focus on the pure 3D structure. Moreover, the nature of amino acids themselves can vary since only part of the residues are used to bind. The only meaningful information is thus the geometric configuration of the amino acids in space.
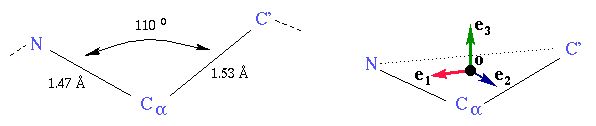
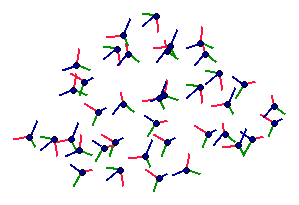
Each amino acid has 4 atoms participating to the backbone of the protein, 3 of them always being in the same geometric configuration. We use these 3 atoms to define the configuration of the amino acid in space. Each amino acid is thus modeled as a frame, and a protein is modeled by an unordered set of frames.
 |
 |
|
Backbone of the protein |
Protein modeled as a set of frames |
To find similar substructures in two proteins, we now have to find two subsets of frames that are in the same configuration, up to a global rigid transformation.